Research
The Kirkland Lab is part of the Cell Cycle & Cancer Biology Research Program at the Oklahoma Medical Research Foundation where we study the role of chromatin regulators in cancer and development
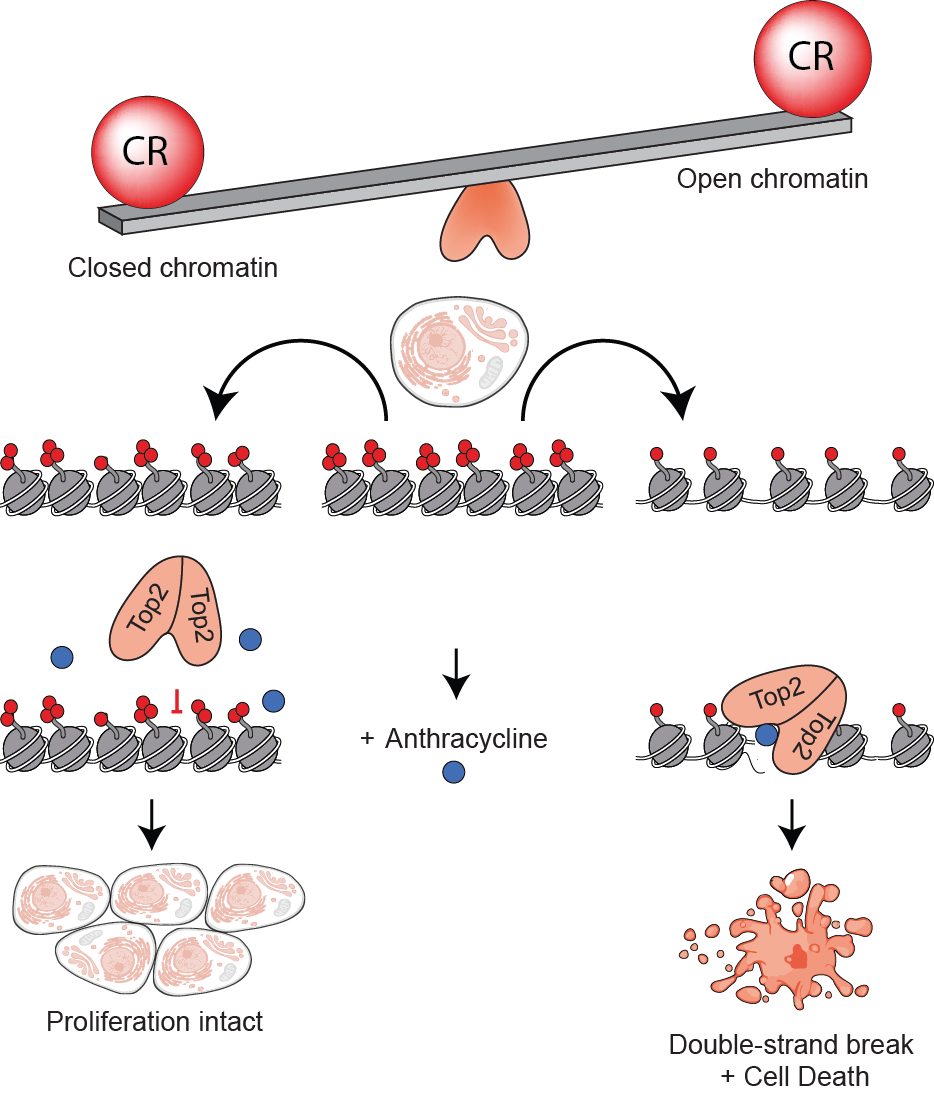
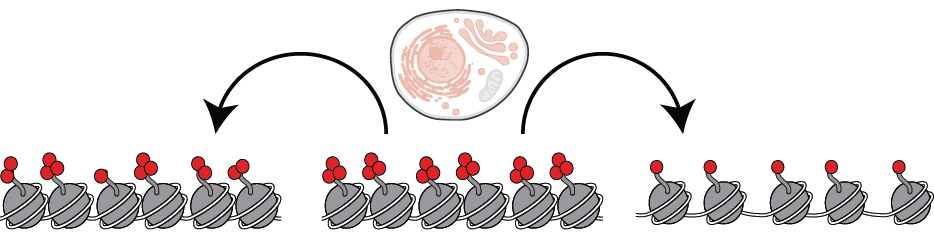
The accessibility of the genome to regulatory, recombination and repair proteins is controlled in part by the opposition between trithorax and polycomb genes that fine-tune accessibility, histone modifications and gene activity. Breast cancer is largely driven by copy-number variation (CNV) and altered gene dosage. What leads to CNV in breast cancer is largely unknown, but CNV may result from structural and organizational changes in the nucleus as a result of altered chromatin regulation. To determine chromatin regulators that may contribute to this phenomenon I collaborated with the lab of Christina Curtis recently published at Nature Medicine. I specifically served as the bridge between the dry and wet lab portions of this study and performed all wet lab experiments. Our results indicate that CRGs operate within a breast cancer specific co-variance transcriptional network that includes members of the classic polycomb and trithorax genes but broadly extends to over 100 chromatin regulators. To validate the extensive chromatin regulatory network, we used a systems biology approach with the aim of achieving a better understanding of chromatin regulator modulation of anthracycline response in breast cancer patients. Since anthracyclines work in part via inhibition of topoisomerase-II (TOP2) on accessible DNA, I hypothesized that CRGs that mediate DNA accessibility might predict anthracycline response. Using cell line datasets and evaluating the interaction between CRG expression and treatment in predicting survival in a metacohort of early-stage breast cancer patients, I identify CRGs whose expression levels dictate anthracycline benefit across the clinical subgroups. Consistent with my hypothesis, CRGs that promote DNA accessibility, including trithorax complex members, were associated with anthracycline sensitivity when highly expressed, whereas CRGs that reduce accessibility, were associated with decreased anthracycline sensitivity. Using molecular biology approaches I validated that expression levels of a CR, KDM4B, modulates TOP2 accessibility to chromatin, elucidating a new pathway of anthracycline resistance. Our novel data-driven methodology identifies a collection of chromatin regulatory genes that form a cancer specific covariate network, parts of which predict response to TOP2 inhibitors likely through alteration of chromatin accessibility. This chromatin regulatory network will inform a robust signature of anthracycline (TOP2i) response that is broadly applicable and provides insight for novel therapeutic targeting and with implications for breast cancer patient stratification and treatment decisions.
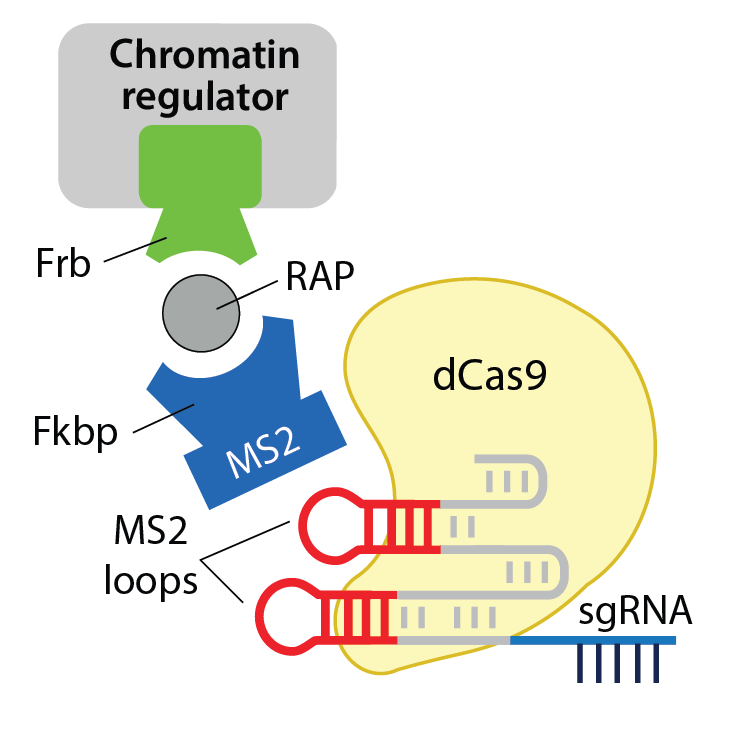
The mSWI/SNF complexes, are mutated in 20% of tumors making them one of the most mutated CR complexes in cancer4. In the past, the mechanism of action of ATP-dependent chromatin regulators, such as mSWI/SNF was largely explored using in vitro methods based on measurements of nucleosome mobility. These studies were limited by the fact that these assays were not sensitive to tissue-specific chromatin modifications, topology, long-range interactions, and complex patterns of histone modifications as well as patterns of DNA methylation and human disease mutational backgrounds. To circumvent these problems I developed the FIRE-Cas9 system that allows one to recruit a specific chromatin regulator and follow the consequences with minute-by-minute kinetics on physiologic chromatin, which I published as a co-first author in Nature Communications. I can examine any locus of biologic or medical importance in virtually any cell type using this technique. Because CR complexes have alterations in many human cancers, it is imperative to understand the mechanism of action of these complexes with unparalleled precision as a basis for therapeutic development. The FIRE-Cas9 system that I developed allows one to do this for the first time.